The DIADIST project is funded by the Biotechnology and Biological Sciences Research Council (BBSRC) under its Bioinformatics Programme (official title: Visual Indexing for Taxonomic Information Systems; grant number 754/BIO14262). It is a collaboration between researchers from Cardiff University, Wales, and the Royal Botanic Garden Edinburgh, Scotland (see staff list for details).
Aim of the project
The aim of this project is to investigate methods to enable the visual
indexing of images and drawings of biological specimens held in a database.
Biological specimens are frequently described in visual form for taxonomic
and other purposes. Vast catalogues of specimen material have been accumulated
over many years in the form of microscope slides, drawings and photographs.
Recently, efforts have been made to digitize such data for electronic storage
and transmission. This research programme will provide a means to structure
taxonomic databases to enable this data to be more effectively queried.
Whilst in some areas of biological data retrieval textual keys may be appropriate, this mode of operation is not adequate where visual data is of major importance, as in taxonomy. A great proportion of microscopic species are described and classified visually for taxonomic purposes. Current biological databases do not exploit the full potential of visual data which may already be stored within them.
There has been some recent work in indexing between images in biological databases [1, 2]. However, there is a clear need to extend such indexing capabilities; the inclusion of biological drawings is a natural extension. The approach taken in the DIADIST project is novel in that it seeks to incorporate taxonomic drawings as a prime source of taxonomic data and to develop methods to enable indexing between digital photographic images and drawings stored in a biological database.
Methodology and Work Programme
We propose to develop a system that will take as input a series of
images and corresponding drawings of selected specimens and will learn
the relationship (mapping) between the images and drawings. A new image
suitably presented to the system can then be mapped into the drawing representation
and classified according to which drawing (type) it most closely matches.
Essentially, the principle used is to seek to transform the "high-dimensional" image space into a "lower-dimensional" space where only relevant features are represented. The mapping essentially extracts the salient image information pertaining to the photograph or drawing (Fig 1).
In our proposed system, corresponding low dimensional representations
of images and drawings are presented, in pairs; by computing a mapping
from an image to a drawing we will train the computer to recognize and
extract the key features needed for taxonomic classification.
We aim to produce a generic system that can operate across a wide range
of biological species. We have selected three taxonomic groups as test
cases for our system: diatoms, desmids, and water mites (Acari).
Recent work at Cardiff [3, 4] has developed methods using extended depth of focus to obtain 3D structure from a series of 2D microscope images serial sections. With the combination of the extended depth of focus method, the image-to-drawing-conversion method, and visual indexing taxonomic drawings will be able to be used to their full potential. One important use of the extended depth of focus method as applied to both diatoms and desmids would be as tools for describing new taxa, both by means of single digital images showing all morphologically relevant detail in focus, and by means of drawings produced automatically on the basis of these images.
Figure 1: Extended depth of focus image of specimen of desmid (top) and of mite (bottom right), standard image of diatom (bottom left), and corresponding drawings
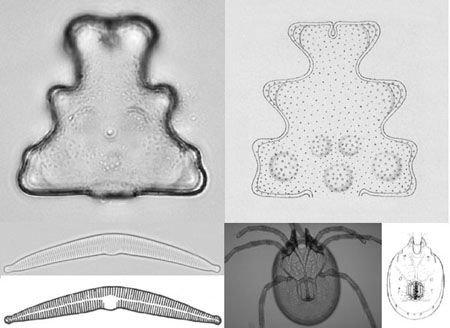
The basic outline of our method is as follows; it has two phases:
Learning phase:
1. Input a set of images and a set of corresponding drawings of a given
set of type specimens, one image per drawing
2. Compute a low-dimensional representation of the images and another
low-dimensional representation of the drawings
3. Compute a mapping between these low-dimensional representations.
Matching phase:
1. Input a new image which is not one of the original images but belongs
to one of the species in the drawings.
2. Represent the image in the low-dimensional way just learned.
3. Map this low-dimensional representation into a new drawing.
4. Find the closest matching type specimen by comparing this to the
known drawings.
Two extensions can readily be developed from the methods above:
Matching between two images
If type data exist as images rather than drawings then the transformation
from image to drawing can be omitted and corresponding low-level image
representations matched directly. Alternatively, both images can be mapped
to drawings and compared.
Producing new drawings
If a new image is presented, a corresponding drawing can be synthesised.
Type drawings are intended to exhibit the general characteristics of a
taxon; however, individual specimens may vary morphologically, and it is
possible that several images could be presented and an average type drawing
assembled. This could be achieved by simply averaging the feature descriptions,
or by using some criteria to select features that best represent the type.
References
1. H. du Buf et al.
Diatom identification: a double challenge called ADIAC. In
Proc. 10th Int. Conf. on Image Analysis and Processing,Venice, Italy,
pages 734-739, September 27-29 1999.
2. P.J.D. Weeks, M.A.
O'Neill, K.J. Gaston, and I.D. Gauld. Species identification of wasps using
principal component associative memories. Image and Vision Computing,
17:861-866, 1999.
3. A.G. Valdecasas, J.M.
Becerra, and A.D. Marshall. Extending the availability of microscope type
material for taxonomy and research. Trends in Ecology and Evolution,
12(6), 1997.
4. A.G. Valdecasas, J.M.
Becerra, and A.D. Marshall. On the extended depth of focus imaging for
bright field microscopy.Micron, 32:
559-569, 2000.
5. C.A. Glasbey and M.G. Perkins. Recovery of depth
information from optical microscope images by constrained deconvolution.
Scandinavian Image Analysis Conference - 97SCIA,
Lappeenranta, Finland, June 1997.